

| Research: Extending Chimera for Cryo-Electron Tomography |


|
|
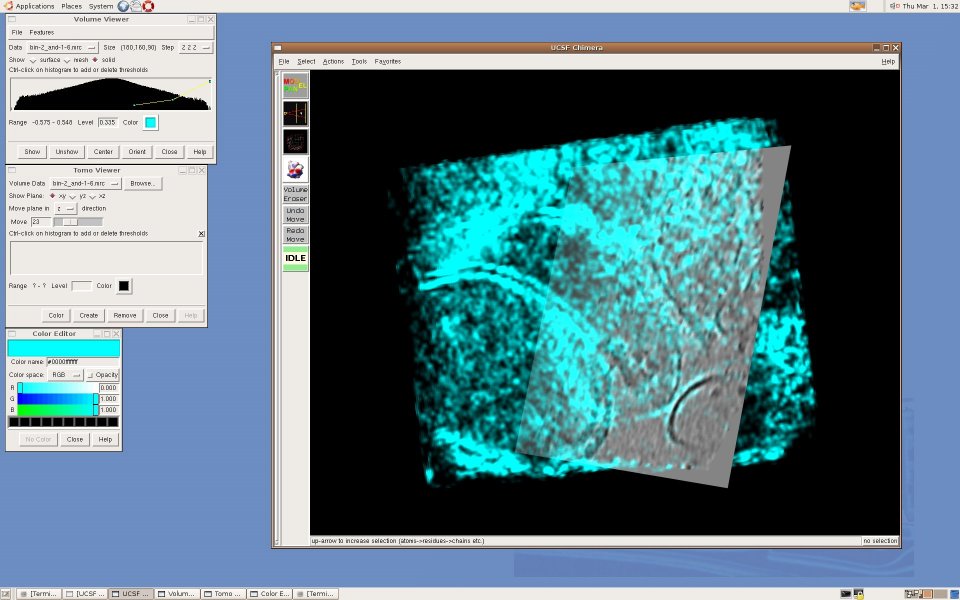
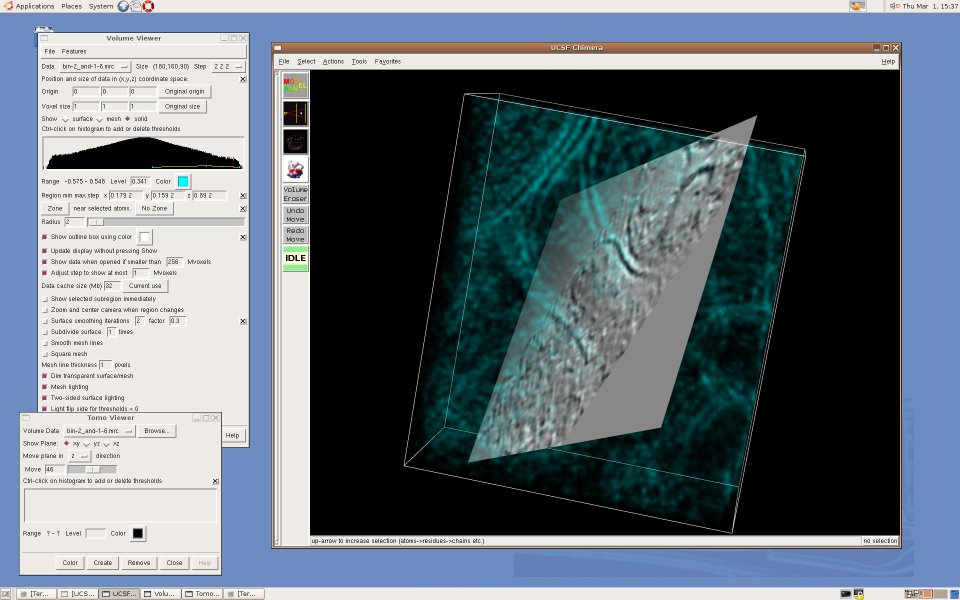
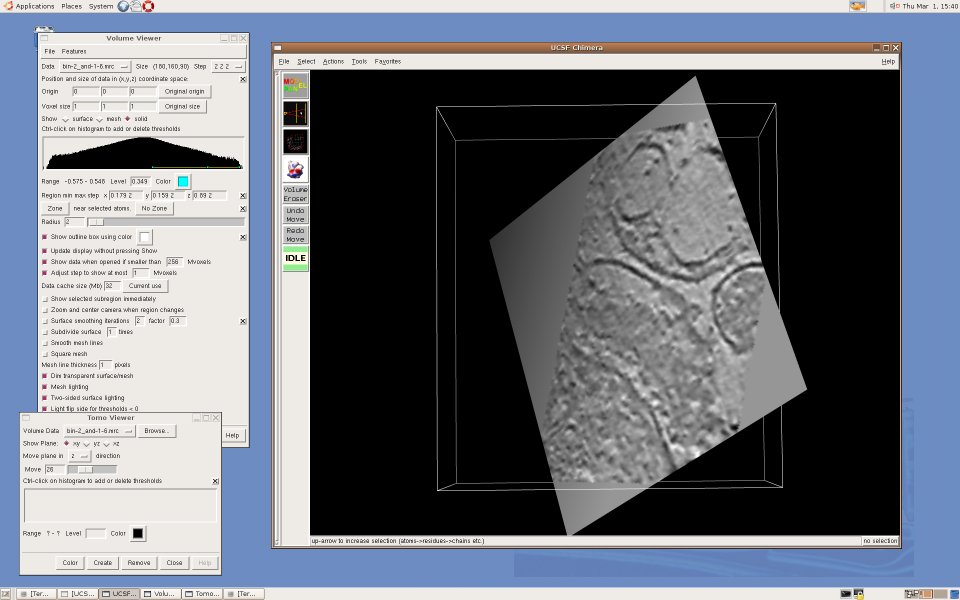
UCSF Chimera is a visualization tool for structural biology developed by the Computer Graphics Laboratory at UC San Francisco. For us, Chimera's appeal is in the fact that it is mostly written in Python and thus easily extensible for exploratory research - for example, to investigate new segmentation algorithms, or try out new ways to visualize electron tomography. Moreover, it is developed by the Resource for Biocomputing, Visualization, and Informatics, funded by NIH's National Center for Research Resources, and thus besides being open-source software, we found that the Chimera team can offer us expertise and support far beyond what we ordinarily get from commercial suppliers. Currently, Karin Gross is developing an extension that creates a "slice" view of a 3D volume, a representation frequently used to visualize highly noisy tomographic data sets. Here are some screenshots: [Cryo-electron tomogram of a rat synapse courtesy of V. Lucic, Martinsried.] An alpha release of TomoPlane can be found in the TomoPlane software page. 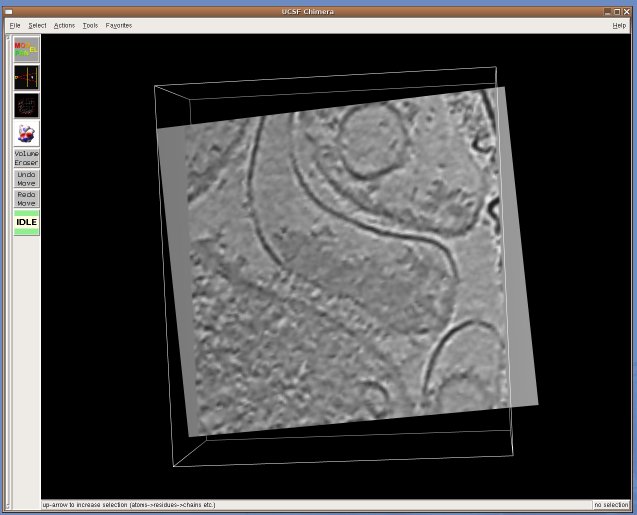
 |
||
|
||